(Originally posted at Planetary Technologies, 04 October, 2019. Written in advance of the 2019 White House Bioeconomy Summit.)
Summary. The end of petroleum is in sight. The reason is simple: the black goo that powered and built the 20th century is now losing economically to other technologies. Petroleum is facing competition at both ends of the barrel, from low value, high volume commodities such as fuel, up through high value, low volume chemicals. Electric vehicles and renewable energy will be the most visible threats to commodity transportation fuel demand in the short term, gradually outcompeting petroleum via both energy efficiency and capital efficiency. Biotechnology will then deliver the coup de grace, first by displacing high value petrochemicals with products that have lower energy and carbon costs, and then by delivering new carbon negative biochemicals and biomaterials that cannot be manufactured easily or economically, if at all, from petrochemical feedstocks.
Bioeconomy Capital is investing to accelerate, and to profit from, the transition away from petroleum to biomanufacturing. We will continue to pursue this strategy beyond the endgame of oil into the coming era when sophisticated biological technologies completely displace petrochemicals, powered by renewable energy and containing only renewable carbon. We place capital with companies that are building critical infrastructure for the 21st century global economy. There is a great foundation to build on.
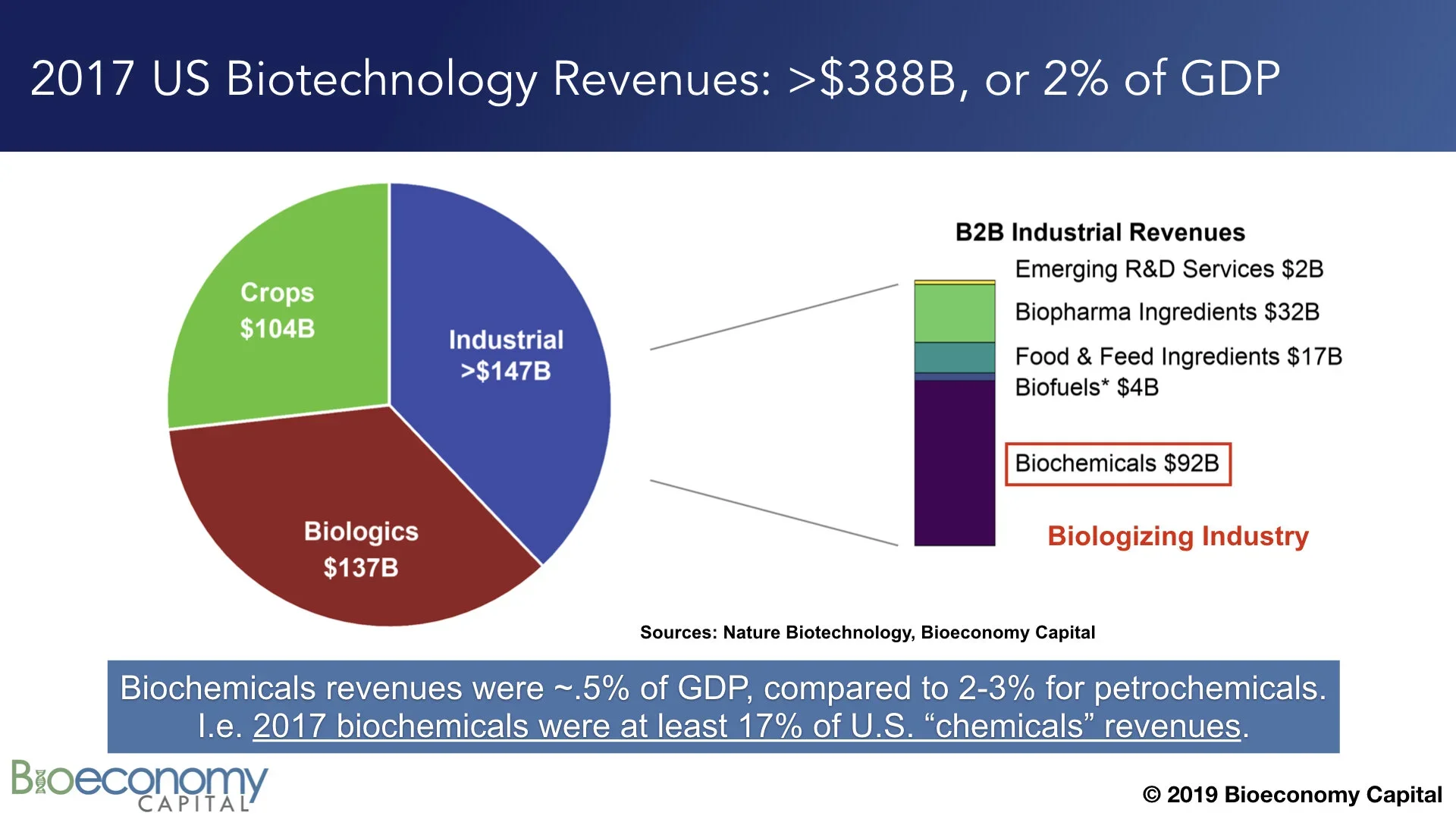
Biotechnology is already an enormous industry in the U.S., contributing more than 2% of GDP (see “Estimating the biotech sector's contribution to the US economy”; updates on the Bioeconomy Dashboard). The largest component of the sector, industrial biotechnology, comprises materials, enzymes, and tools, with biochemicals alone generating nearly $100B in revenues in 2017 (note that this figure excludes biofuels). That $100B is already between 1/6 and 1/4 of fine chemicals revenues in the U.S., depending on whether you prefer to use data from industrial associations or from the government. In other words, biochemicals are already outcompeting petrochemicals in some categories. That displacement is a clear indication that the global economy is well into shifting away from petrochemicals.
See the Bioeconomy Dashboard for downloadable graphics and additional analysis.
The common pushback to any story about the end of fossil fuels is to assert that nothing can be cheaper than an energy-rich resource that oozes from a hole in the ground. But, as we shall see, that claim is now simply, demonstrably, false for most petroleum production and refining, particularly when you include the capital required to deliver the end use of that petroleum. It is true that raw petroleum is energy rich. But it is also true that it takes a great deal of energy, and a great deal of capital-intensive infrastructure, to process and separate oil into useful components. Those components have quite different economic value depending on their uses. And it is through examining the economics of those different uses that one can see the end of oil coming.
First, let us be clear: the demise of the petroleum industry as we know it will not come suddenly. Oil became a critical energy and materials feedstock for the global economy over more than a century, and oil is not going to disappear overnight. Nor will the transition be smooth. Revenues from oil are today integral to maintaining many national budgets, and thus governments, around the globe. As oil fades away, governments that continue to rely on petroleum revenues will be forced to reduce spending. Those governments have a relatively brief window to diversify their economies away from heavy reliance on oil, for example by investing in domestic development of biotechnology. Without that diversification, some of those governments may fall because they cannot pay their bills. Yet even when oil’s clear decline becomes apparent to everyone, it will linger for many years. Government revenues for low cost producers (e.g., Iran and Saudi Arabia) will last longer than high cost producers (e.g. Brazil and Canada). But the end is coming, and it will be delivered by the interaction of many different technical and economic trends. This post is an outline of how all the parts will come together.
WHAT PRODUCES VALUE IN A BARREL OF OIL? ERGS AND ATOMS
Any analysis of the future of petroleum that purports to make sense of the industry must grapple with two kinds of complexity. Firstly, the industry as a whole is enormously complex, with different economic factors at work in different geographies and subsectors, and with those subsectors in turn relying on a wide variety of technologies and processes. In 2017, The Economist published a useful graphic (below — click through to the original) and story (“The world in a barrel”) that explored this complexity. Moreover, the cost of recovering a barrel and delivering it to market in different countries varies widely, between $10 and $70. Further complicating analysis, those reported cost estimates also vary widely, depending on both the data source and the analyst: here is The Economist, and here is the WSJ, and note that these articles cite the same source data but report quite different costs. The total market value of petroleum products is about $2T per year, a figure that of course varies with the price of crude.
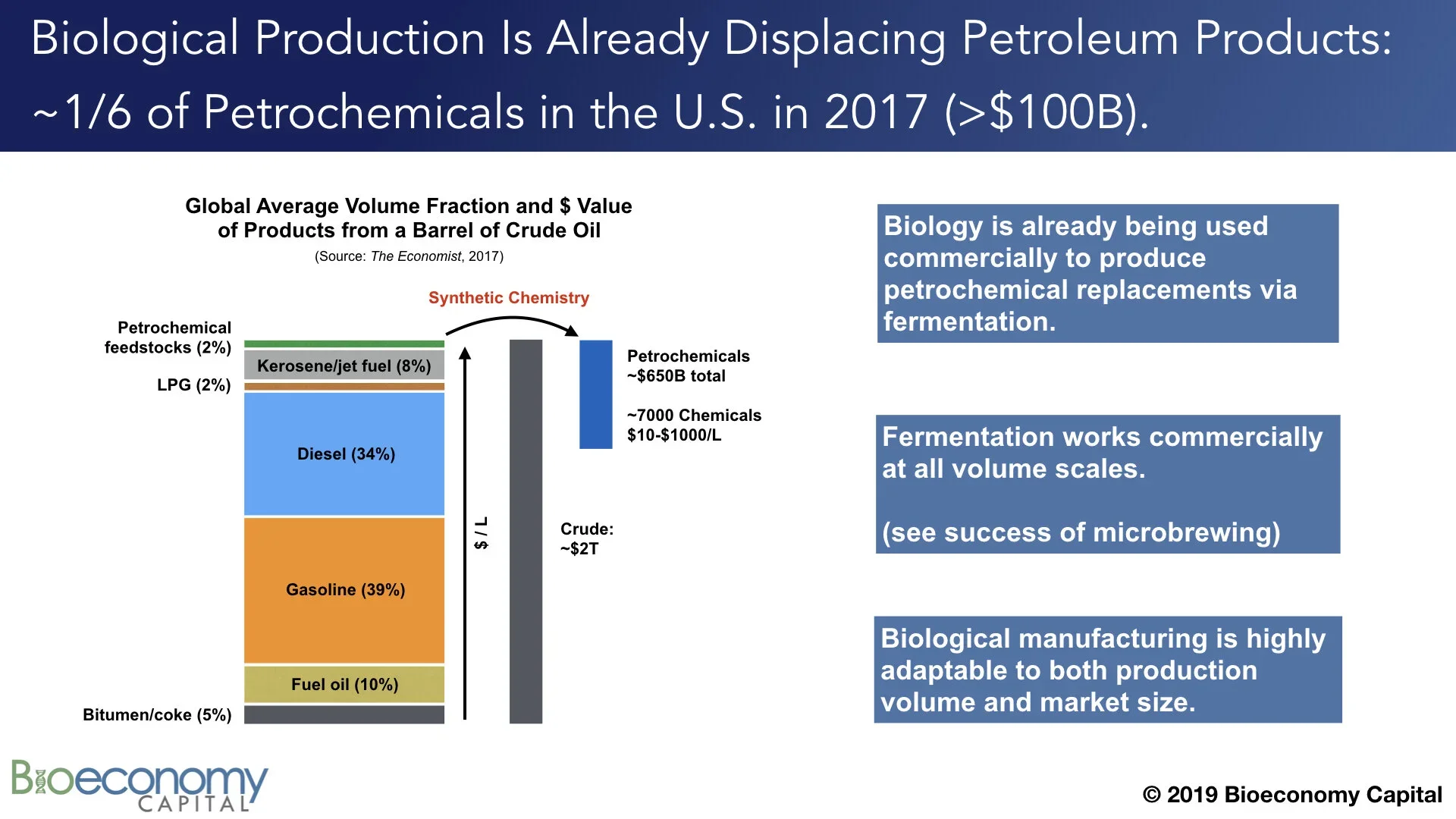
Secondly, “a barrel of oil” is itself complex; that is, barrels are neither the same nor internally homogeneous. Not only are barrels from different wells composed of different spectra of molecules (see the lower left panel above in “Breaking down oil”), but those molecules have very different end uses. Notably, on average, of the approximately 44 gallons per barrel worth of products that are generated during petroleum refining, >90% winds up as heating oil or transportation fuel. Another approximately 5% comprises bitumen and coke. Both of these are are low value; bitumen (aka “tar”) gets put on roads and coke is often combined with coal and burned. In other words, about 42 of the 44 gallons of products from a barrel of oil are applied to roads or burned for the energy (the ergs) they contain.
The other 2% of a barrel, or 1-2 gallons depending on where it comes from, comprise the matter (the atoms) from which we build our world today. This includes plastics precursors, lubricants, solvents, aromatic compounds, and other chemical feedstocks. After being further processed via synthetic chemistry into more complex compounds, these feedstocks wind up as constituents of nearly everything we build and buy. It is widely repeated that chemical products are components of 96% of U.S.-manufactured goods. That small volume fraction of a barrel of oil is thus enormously important for the global economy; just ~2% of the barrel produces ~25% of the final economic value of the original barrel of crude oil, to the tune of more than $650B annually.
CHEAPER ERGS
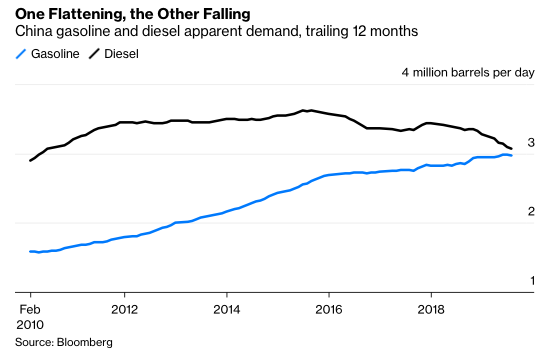
The big news about the ergs in every barrel is that their utility is coming to an end because the internal combustion engine (ICE) is on its way out. Electric vehicles (EVs) are coming in droves. EVs are far more efficient, and have many fewer parts, than ICE powered vehicles. Consequently, maintenance and operating costs for EVs are signficantly lower than for ICE vehicles. Even a relatively expensive Tesla Model 3 is cheaper to own and operate over 15 years than is a Honda Accord. Madly chasing Tesla into the EV market, and somewhat late to the game, Volkswagen has announced it is getting out of manufacturing ICEs altogether. Daimler will invest no more in ICE engineering and will produce only electric cars in the future. Daimler is also launching an electric semi truck in an effort to compete with Tesla’s forthcoming freight hauler. Not to be left out, VW just announced its own large investment into electic semi trucks. Adding to the trend, last week Amazon ordered 100,000 electric delivery trucks. Mass transit is also shifting to EVs. Bloomberg reported earlier in 2019 that, by the end of this year “a cumulative 270,000 barrels a day of diesel demand will have been displaced by electric buses.” In China total diesel demand is already falling, gasoline demand may well peak this year (see below). Bloomberg points to EVs as the culprit. Finally, as described in a recent report by Mark Lewis at BNP Paribas, the combination of renewable electricity and EVs is already 6-7X more capital efficient than fossil fuels and ICEs at delivering you to your destination; i.e. oil would have to fall to $10-$20/barrel to be competitive.
Consequently, for the ~75% of the average barrel already directly facing competition from cheaper electricity provided by renewables, the transition away from oil is already well underway. Gregor Macdonald covers much of this ground quite well in his short book Oil Fall, as well as in his newsletter. Macdonald also demonstrates that renewable electricity generation is growing much faster than is EV deployment, which puts any electricity supply concerns to rest. We can roll out EVs as fast as we can build them, and anyone who buys and drives one will save money compared to owning and operating a new ICE vehicle. Forbes put it succinctly: “Economics of Electric Vehicles Mean Oil's Days As A Transport Fuel Are Numbered.”
But it isn’t just the liquid transportation fuel use of oil that is at risk, because it isn’t just ergs that generate value from oil. Here is where the interlocking bits of the so-called “integrated petroleum industry” are going to cause financial problems. Recall that each barrel of oil is complex, composed of many different volume fractions, which have different values, and which can only be separated via refining. You cannot pick and choose which volume fraction to pull out of the ground. As described above, a disproportionate fraction of the final value of a barrel of oil is due to petrochemicals. In order to get a hold of the 2% of a barrel that constitutes petrochemical feedstocks, and thereby produce the 25% of total value derived from those compounds, you have to extract and handle the other 98% of the barrel. And if you are making less money off that 98% due to decreased demand, then the cost of production for the 2% increases. It is possible to interconvert some of the components of a barrel via cracking and synthesis, which might enable lower value compounds to become higher value compounds, but it is also quite expensive and energy intensive to do so. Worse for the petroleum industry, natural gas can be converted into several low cost petrochemical feedstocks, adding to the competitive headwinds the oil industry will face over the coming decade. Still, there is a broad swath of economically and technologically important petroleum compounds that currently have no obvious replacement. So the real question that we have to answer is not what might displace the ergs in a barrel of oil — that is obvious and already happening via electrification. The much harder question is: where do we get all the complex compounds — that is, the atoms, in the form of petrochemicals and feedstocks — from which we currently build our complex economy? The answer is biology.
Biochemicals are already competing with petrochemicals in a ~$650B global market.
RENEWABLE ATOMS
Bioeconomy Fund 1 portfolio companies Arzeda, Synthace, and Zymergen have already demonstrated that they can design, construct, and optimize new metabolic pathways to directly manufacture any molecule derived from a barrel of oil. Again, at least 17%, and possibly as much as 25%, of US fine chemicals revenues are already generated by products of biotechnology. To be sure, there is considerable work to do before biotechnology can capture the entire ~$650B petrochemical revenue stack. We have to build lots of organisms, and lots of manufacturing capacity in which to grow those organisms. But scores of start-ups and Fortune 50 companies alike are pursuing this goal. As metabolic engineering and biomanufacturing matures, an increasing number of these companies will succeed.
The attraction is obvious: the prices for high value petrochemicals are in the range of $10 to $1000 per liter. And whereas the marginal cost of production for petroleum products is around $20 billion dollars — the cost of a new refinery — the marginal cost of production for biological production looks like a beer brewery, which comes in at between $100,000 and $10 million, depending on the scale. This points to one of the drivers for adopting biotechnology that isn’t yet on the radar for most analysts and investors: the return on capital for biological production will be much higher than for petroleum products, while the risk will be much lower. This gap in understanding the current and future advantages of biology in chemicals manufacturing shows up in overoptimistic growth predictions all across the petroleum industry.
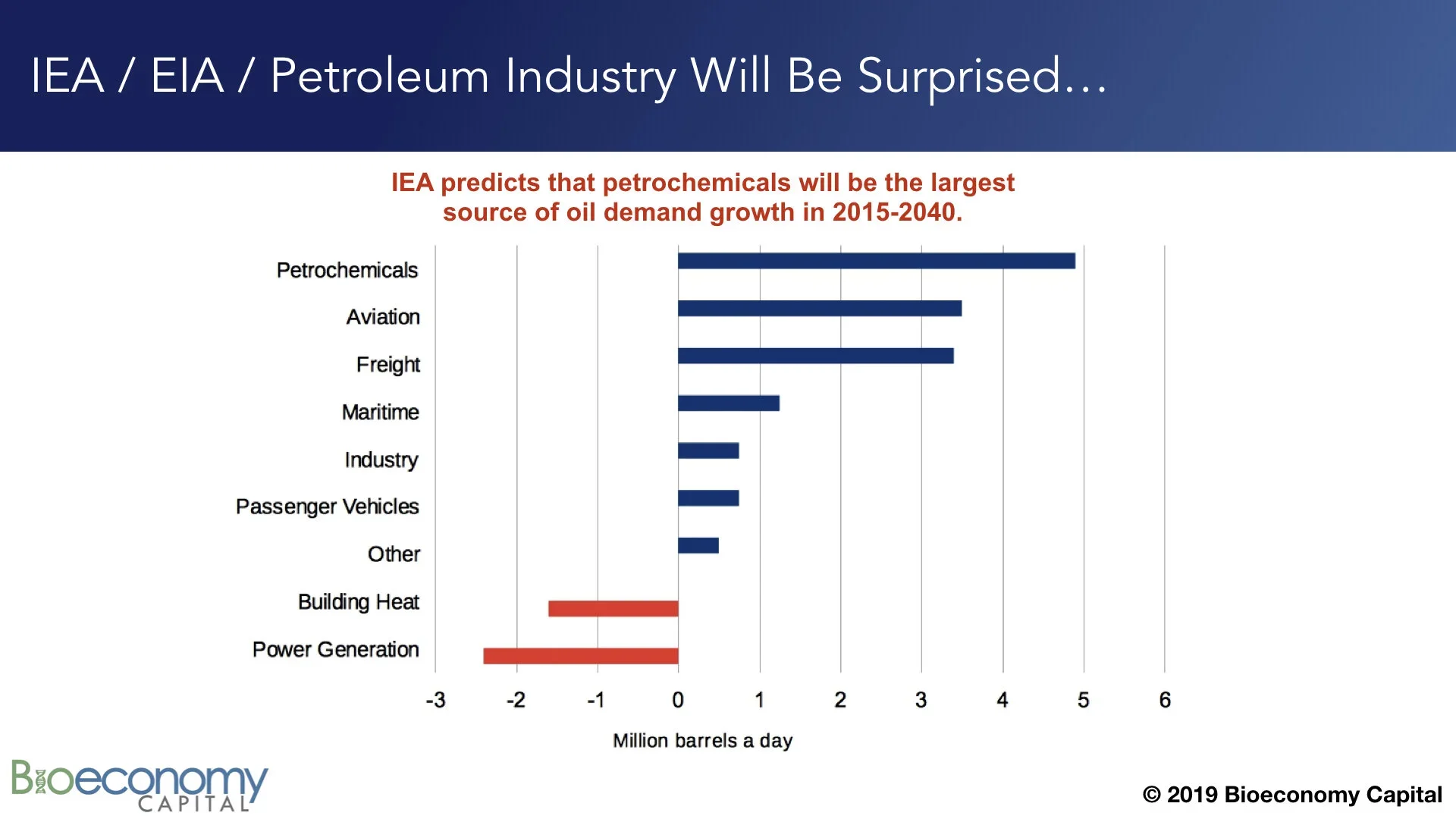
For example, the IEA recently forecast that petrochemicals will account for the largest share of demand growth for the petroleum industry over the next two decades. But the IEA, and the petroleum industry, are likely to be surprised and disappointed by the performance of petrochemicals. This volume fraction is, as noted above, already being replaced by the products of biotechnology. (Expected demand growth in “Passenger vehicles”, “Freight”, and “Industry”, which uses largely comprise transportation fuel and lubricants, will also be disappointing due to electrification.) We should certainly expect the demand for materials to grow, but Bioeconomy Capital is forecasting that by 2030 the bulk of new chemical supply will be provided by biology, and that by 2040 biochemicals will be outcompeting petrochemicals all across the spectrum. This transition could happen faster, depending on how much investment is directed at accelerating the roll out of biological engineering and manufacturing.
Before moving on, we have to address the role of biofuels in the future economy. Because biofuels are very similar to petroleum both technologically and economically — that is, biofuels are high volume, low margin commodities that are burned at low efficiency — they will generally suffer the same fate, and from the same competition, as petroleum. The probable exception is aviation fuel, and perhaps maritime fuel, which may be hard to replace with batteries and electricity for long haul flights and transoceanic surface shipment.
But this likely fate for biofuels points to the use of those atoms in other ways. As of 2019, approximately 10% of U.S. gasoline consumption is contributed by ethanol, as mandated in the Renewable Fuels Standard. That volume is the equivalent of 4% of a barrel of oil, and it is derived from corn kernels. As ethanol demand falls, those renewably-sourced atoms will be useful as feedstocks for products that displace other components of a barrel of oil. The obvious use for those atoms is in the biological manufacture of chemicals. Based on current yields of corn, and ongoing improvements in using more of each corn plant as feedstock, there are more than enough atoms available today just from U.S. corn harvests, let alone other crops, to displace the entire matter stream from oil now used as petrochemical feedstocks.
BEYOND PETROCHEMISTRY
The economic impact of biochemical manufacturing is thus likely to grow significantly over the next decade. Government and private sector investments have resulted in the capability today to biomanufacture not just every molecule that we now derive from a barrel of petroleum, but, using the extraordinary power of protein engineering and metabolic engineering, to also biomanufacture a wide range of new and desirable molecules that cannot plausibly be made using existing chemical engineering techniques. This story is not simply about sustainability. Instead, the power of biology can be used to imbue products with improved properties. There is enormous economic and technical potential here. The resulting new materials, manufactured using biology, will impact a wide range of industries and products, far beyond what has been traditionally considered the purview of biotechnology.
For example, Arzeda is now scaling up the biomanufacturing of a methacrylate compound that can be used to dramatically improve the properties of plexiglass. This compound has long been known by materials scientists, and long been desired by chemical engineers for its utility in improving such properties as temperature resistance and hardness, but no one could figure out how to make it economically in large quantities. Arzeda's biological engineers combined enzymes from different organisms with enzymes that they themselves designed, and that have never existed before, to produce the compound at scale. This new material will shortly find its way into such products as windshields, impact resistant glass, and aircraft canopies.
Similarly, Zymergen is pursuing remarkable new materials that will transform consumer electronics. Zymergen is developing a set of films and coatings that have a set of properties unachievable through synthetic chemistry and that will be used to produce flexible electronics and displays. These materials simply cannot be made using the existing toolbox of synthetic chemistry; biological engineering gives access to a combination of material properties that cannot be formulated any other way. Biological engineering will bring about a renaissance in materials innovation. Petroleum was the foundation of the technology that built the 20th century. Biology is the technology of the 21st century.
FINANCING RISK
The power and flexibility of biological manufacturing create capabilities that the petroleum industry cannot match. Ultimately, however, the petroleum industry will fade away not because demand for energy and materials suddenly disappears, or because that demand is suddenly met by renewable energy and biological manufacturing. Instead, long before competition to supply ergs and atoms displaces the contents of the barrel, petroleum will die by the hand of finance.
The fact that both ends of the barrel are facing competition from technologically and economically superior alternatives will eventually lead to concerns about oil industry revenues. And that concern will reduce enthusiasm for investment. That investment will falter not because total petroleum volumes see an obvious absolute drop, but rather because the contents of the “marginal barrel” – that is, the next barrel produced – will start to be displaced by electricity and by biology. This is already happening in China and in California, as documented by Bloomberg and by Gregor Macdonald. Thus the first sign of danger for the oil industry is that expected growth will not materialize. Because it is growth prospects that typically keep equities prices high via demand for those equities, no growth will lead to low demand, which will lead to falling stock prices. Eventually, the petroleum industry will fail because it stops making money for investors.
The initial signs of that end are already apparent. In an opinion piece in the LA Times, Jagdeep Singh Bachher, the University of California’s chief investment officer and treasurer, and Richard Sherman, chairman of the UC Board of Regents’ Investments Committee, write that “UC investments are going fossil free. But not exactly for the reasons you may think.” Bachher and Sherman made this decision not based on any story about saving the planet or on reducing carbon emissions. The reason for getting rid of these assets, put simply, is that fossil fuels are no longer a good long-term investment, and that other choices will provide better returns:
We believe hanging on to fossil fuel assets is a financial risk [and that] there are more attractive investment opportunities in new energy sources than in old fossil fuels.
An intriguing case study of perceived value and risk is the 3 year saga of the any-day-now-no-really Saudi Aramco IPO. Among the justifications frequently mooted for the IPO is the need to diversify the country's economy away from oil into industries with a brighter future, including biotechnology, that is, to ameliorate risk:
The listing of the company is at the heart of Prince Mohammed’s ambitious plans to revamp the kingdom’s economy, with tens of billions of dollars urgently needed to fund megaprojects and develop new industries.
There have been a few hiccups with this plan. The challenges that Saudi Aramco is facing in its stock market float are multifold, from physical vulnerability to terrorism, to public perception and industry divestment, through to concerns about the long-term price of oil:
When Saudi Arabia’s officials outlined plans to restore output to maximum capacity after attacks that set two major oil facilities ablaze on Saturday, they were also tasked with convincing the world that the national oil company Saudi Aramco was investable.
The notion that the largest petroleum company in the world might have trouble justifying its IPO, and might have trouble hitting the valuation necessary to raise the cash its current owners are looking for, is eye opening. This uncertainty creates the impression that Aramco may have left it too late. The Company managers may see less value from their assets than they had hoped, precisely because increased financial risk is reducing that value.
And that is the point — each of the factors discussed in this post increases the financing risk for the petroleum industry. Risk increases the cost of capital, and when financiers find better returns elsewhere they rapidly exit the scene. This story will play out for petroleum investments just as it has for coal. Watch what the bankers do; they don’t like to lose money, and the writing is on the wall already. In 2018, global investment in renewable electricity generation was three times larger than the investment in fossil fuel powered generation. Biotechnology already provides at least 17% of chemical industry revenues in the U.S., and is growing in the range of 10-20% annually (see the inset in Figure 2). If you put the pieces together, you can already see the end of oil coming.